
-logo.png)






UV-Vis spectroscopic measurements consist one of the most widely used methods for protein quantification. This is due to the fact that the tryptophan and tyrosine (and to a lesser extent the phenylalanine) residues of proteins strongly absorb at 280 nm. Hence, the corresponding absorbance is used for the calculation of protein concentration, based on specific quantification formulas. If the protein sample does not have tryptophan or tyrosine, the concentration can still be easily measured by the absorption at 205 nm in which the peptide bonds are analysed directly (Scopes method). Conformational studies can also be performed under special conditions.
The spectroscopic equipment of TPCI/NHRF accessible through the Instruct-EL hub includes a UV-Vis-NIR spectrophotometer (Lambda 19 by Perkin Elmer), which is a general-purpose instrument for absorption spectroscopy in the ultraviolet, visible, and near infrared spectral regions. It allows measurement of transmittance and reflectance of diffuse or scattering materials as a function of wavelength. Moreover, the optical scheme consists of a double beam double monochromator configuration, while the useable wavelength range is 250 – 2500 nm with temperature control. UV-Vis spectroscopic services are mainly focused towards protein quantification and concentration determination. Moreover, reflectance measurements of thin films and surfaces is also available.
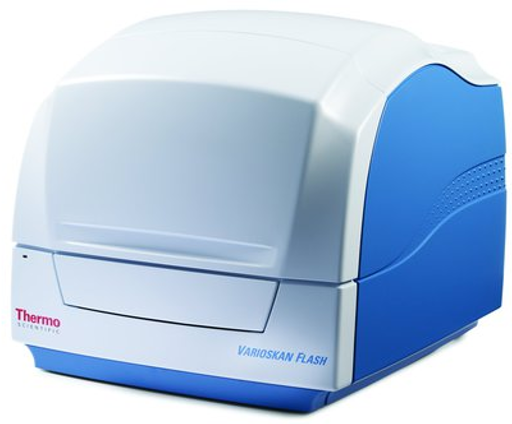
DESCRIPTION: Microplate reader Vis/UV/Fluorescence/ Luminescence
FUNDER: FP7-REGPOT-2009-1, No245866, "ARCADE"

DESCRIPTION: Protein structure determination and studies on physicochemical properties in protein aqueous solutions
FUNDER: General Secretariat for Research and Technology
A double beam UV-Vis spectrophotometer (HITACHI U-2001) is available in the HPI’s Bacteriology laboratory measuring absorbance at the range 190 – 1100 nm with temperatures being controlled trough a water circulation apparatus. The device can be used for a variety of applications such as protein and nucleic acid quantification, wave length scans, and time scans permitting the real time monitoring of enzymatic reactions for steady state kinetic analyses and inhibition studies. The instrument is PC controlled and the relevant analyses can be performed with the UV Visions software (Hitachi).

DESCRIPTION:
FUNDER:
Visit the provider's website: http://inspired.aua.gr/instruct/

DESCRIPTION:
FUNDER: INSPIRED-AUA GSRT

DESCRIPTION:
Basic Functions:
Photometry: Test Abs., Transmittance and Energy by fixed wavelength
Quantitative: Linear fit and Linear fit through zero two modes
Coefficient, Standard Sample input and Standard Sample read three modes to establish standard curve
Establish A=K1*C+K0, can search original data, graph curve, parameters settings
Can save 240 group curves, can test 240 data in each curve
Double wavelength, Triple wavelength test functions
Kinetics: Used for time course scanning or reaction rate calculations△A/t, can search all data.
Multi-Wavelength: Can test Transmittance and Abs. with 8 different wavelengths at most
Scanning: User can set the scan range from 190nm to 1100nm to test the max. Abs. peak value,
can do derivation, arithmetical operations to the graph.
Biology: DNA/Protein, (Optional: UV, Lowry, BCA, CBB and Biuret)
FUNDER: IKY (State Scholarships Foundation) Fellowships of Excellence for Postgraduate Studies in Greece-Siemens Program.
DESCRIPTION:
FUNDER:
The Hitachi's U-5100 UV-Visible Spectrophotometer is a general-purpose instrument for absorption spectroscopy in the ultraviolet, visible, and near infrared spectral regions. It delivers a compact, lightweight package with remarkable power savings and a long life for its light source. The optical scheme consists of a Seya-Namioka mount monochromator configuration and the useable wavelength range is 190-1100 nm with temperature control. UV-Vis spectroscopic services are mainly focused towards protein quantification and concentration determination. Moreover, It contains automatic switching of 6 cells by using an automatic 6-cell turret and a high-speed scanning (12,000 nm/min) deliver a substantial reduction in measurement time (approximately 60% reduction).

DESCRIPTION:
FUNDER:
This is some info